Summary
Gene editing has made a lot of progress over the past few decades. CRISPR-Cas9 sparked the recent fervor and won some of its early pioneers a Nobel Prize, and more recently Prime Editing and Cytosine/Adenine Base Editors (CBEs/ABEs) have further expanded the range of editing targets and types of edits that are possible while increasing efficiency and reducing the rate of off-target effects.
From the perspective of safe, ubiquitous editing in humans using these systems, a few major challenges for editing remain towards translating novel target discovery into clinical application:
Off-target edits: All editors still sometimes perform unwanted (off-target) edits. These edits can be close to the target edit site (proximal) or in totally different regions of the genome (distal). As we’ll discuss more, it’s hard to pin down exactly how much of a problem these edits are relative to normal DNA damage (which happens constantly) because the rates haven’t been benchmarked against each other, but our current sense is that they’re high enough to be a bottleneck for using editing for common, non-life threatening diseases or to modify traits.
Immunogenicity: Compared to off-targeting concerns, immunogenicity is a much more nebulous consideration when thinking about the future of gene editing. It is certainly clear that immune responses from vectors is a problem, but there are open ended questions about whether reactions to CRISPR effectors themselves should delay clinical progress. Largely, we feel it’s less of a bottleneck.
Universality: Fixing many concurrent problems in the genome requires being able to land editing constructs freely. Because of the way various editing mechanisms work, not every part of the genome is accessible to editing. However, lots of great work has happened to allow rather uninhibited access to whatever target scientists feel are important to go after. Of all of the concerns with repeated edits, this seems the closest to solved.
We discuss this more in Future directions and projects we’re excited about but at a high level we see 1) larger, open datasets designed for use by ML and other computational methods and 2) longitudinal/observational studies of the impact of edits in vivo as two of the most promising and neglected buckets for accelerating field-wide progress.
Zooming out, other major challenges remain: deciding what changes to make, figuring out a regulatory regime that can accommodate such a powerful technology, scaling therapeutics up, and many more– all interesting problems in their own right, but not problems we will touch on in this piece.
Motivation
People have an interesting (to us) attitude towards genetic inequity. They often just accept that some people only need 4-6 hours of sleep with seemingly no downsides (even accounting for pleiotropy) besides hyperactivity, others are nearly immune to Alzheimer's or heart disease, or use oxygen much more efficiently, among many other examples and even bristle at the idea that we should make these traits more widely available. While we agree it’s important to discuss the implications, we see the kneejerk anti- response as a status quo bias, especially since soon things won’t have to be this way.
With the current rate of progress, it seems quite likely that in the next two decades, small somatic edits for common diseases and even trait modification may be possible. Right now, most research understandably focuses on fixing a single problem with a single administration per person. However, if our ultimate goal is control over our own genes, we need to look ahead to what will stop us from getting there.
Why the focus on gene editing instead of other methods of improving health or changing traits, like traditional drug pharmacology? One key difference is genetic editing would theoretically be modular in a way that designing a new small molecule would not be; once a reliable process for editing a specific tissue is identified, different edits would require changes in the template DNA or RNA, but not redesign of the entire drug delivery system. Secondly, the presence of existing humans with various mutations effectively serves as (free) natural sources of discovery for what types of modifications are beneficial, and which are deleterious. Three, editing provides a much wider range of options for how we can reach our goals. Small molecules typically (although not always) operate by binding a protein thereby preventing its uptake or performing some form of inhibition. Editing, on the other hand, allows us to apply the full range of genetic interventions (knock-outs, upregulation, modification). Put a different way, genetic medicine ultimately succeeds where drug targeting fails because they target the exact things that are broken within us. All therapies start at a known mechanism, and work their way into technical feasibility. Most importantly, gene therapy can cure disease for a lifetime – something that makes it stand much taller than its small molecule peers.
This post outlines what we know works well about the tools at our disposal in our quest for safe somatic gene editing, and highlights areas that could benefit from more work, especially work that might be neglected by academia and industry. Mainly, this means focusing on what specific gaps prevent repeated, targeted delivery of engineered Cas constructs for non-life threatening diseases and trait modification. For simplicity we will define the problem with respect to small substitutions and deletions, leaving larger insertions for another post (though we want to acknowledge that a ton of incredible things are happening in this area right now).
Delivery
As mentioned in the introduction, any in vivo gene edits require a way of getting the editing components into the cell. There are numerous delivery vectors available, each with its own set of advantages and drawbacks. While we won't delve into the specifics of each vector in this post, we recommend checking out our blog posts on this topic for a more in-depth exploration. In the context of repeatable, safe gene editing, there are three primary challenges we must address: efficiency, immunogenicity, and packaging size.
Efficiency and Specificity
A significant concern in gene editing is the ability to target specific tissues or even cell types while avoiding others. In many cases, we want to edit only certain tissues to minimize potential side effects and toxicity. Additionally, organs like the liver might take up too much vector, posing an additional risk of toxicity.
For multiplex editing, the challenge is even greater. The amount of vector needed typically increases linearly with the number of distinct edits required, for example to accommodate multiple guide RNAs. This makes it more difficult to achieve efficient and specific gene editing compared to single-gene therapies.
Immunogenicity of Vectors
In some instances, trading off efficiency for the ability to redose might be acceptable. However, redosing poses the risk of provoking an immune response. The immune system can react to both the vector and the editor, but in this context, we are primarily concerned with the vector’s immunogenicity. For a more detailed discussion on the challenges specific to adeno-associated virus (AAV) vectors and immunogenicity, see here.
Vector Packaging Size
Figure source: Engineering Gene Therapy: Advances and Barriers. Note: more recent engineered Adenovirus variants can actually package up to 36kb payloads.
Figure source: Viral Delivery Systems for CRISPR
Different viral (and nonviral) vectors have very different packaging capacities, which trade off against other factors such as immunogenicity and efficiency. When we talk about packaging size here, we’re specifically focused on delivering nucleotides. Delivering proteins or other biological material directly could also be valuable, but we’re ignoring it in this section. Different variants of the Cas protein have genomes ranging in sizes of 3-5kb. Once you add in a gRNA, all the other gene regulatory machinery, and other miscellaneous genetic elements, the full therapeutic construct typically takes up at least 5kb. This means it’s larger than what a wild type AAV can package. Since AAV is attractive for other reasons, researchers have explored various strategies to reduce the size of the editing components, such as deleting amino acids, developing mini-Cas9 systems, or utilizing Cas9 with polyethylene glycol (PEG) modifications.
Editing
The Landscape and Background of Gene Editing Technologies
Even with great delivery systems, editing still remains a major barrier to repeated edits. The core technology has improved tremendously over the last few years, with several key technologies leading the way.
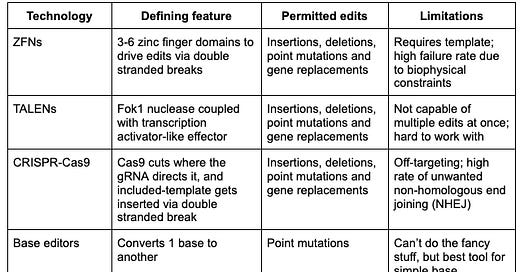
Summary table
For those already in the know, or particularly impatient readers looking to save a couple minutes.
Pre-CRISPR editors
ZFNs (Zinc Finger Nucleases)
ZFNs are artificial restriction enzymes that combine a zinc finger DNA-binding domain with a FokI nuclease domain. Though the specific number varies, the typical ZFN makes use of between three to six three-base pair recognition zinc finger domains, which then work in complex to recognize an entire target gene. Once a targeted double stranded break is made by the cleavage domain, gene replacement can be done, albeit through natural cellular repair mechanisms. While recent ML-guided design of ZFNs may catalyze a renaissance, ZFNs currently play an insignificant role in the broader editing landscape because of the high failure rate resulting from biophysical constraints of the modular assembly method.
TALENs (Transcription Activator-Like Effector Nucleases)
TALENs are customizable nucleases that combine a transcription activator-like effector (TALE) DNA-binding domain and a FokI nuclease (N) domain. The DNA-binding domain is composed of tandem repeats, each of which recognizes a single base pair in the target DNA sequence. The specificity of TALENs can be enhanced by designing longer DNA-binding domains, which reduces the likelihood of off-target binding. Though still sometimes found in niche basic science research applications where multiple edits – which are possible with CRISPR systems – are not needed, TALENs has largely fallen out of favor in the editing space in recent years due to its technical limitations, namely the inability to target multiple sites and complexity in designing the constructs themselves.
New era editors
CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats and CRISPR-associated protein 9)
Figure source: UMass RNA Therapeutics Institute
CRISPR-Cas9 is a versatile gene editing system that behaves like a molecular scissor tailor-made for a specific gene of interest. Genome editing is carried out via the Type II CRISPR-Cas9 system, which can be generalized into three main parts: a single guide RNA (gRNA), a Cas9 nuclease, and a repair template. First, the gRNA recognizes and directs the CRISPR-system, which includes the Cas9 nuclease itself, to the gene of interest. A critical piece of the CRISPR-Cas9 system is the presence of a protospacer adjacent motif (PAM) directly adjacent to the targeted gene in the host genome, which serves as the recognition site for the Cas9 nuclease itself. Once bound to the 20 bp PAM sequence, Cas9 begins to introduce double-stranded breaks, resulting in two possible fates: homology-directed repair (HDR – good!) and non-homologous end joining (NHEJ – bad!). HDR utilizes exogenous DNA included in the CRISPR construct to serve as a template for repair once a double stranded break is recognized by the cell’s natural machinery. In the cases where HDR is not possible, the cell simply fixes the gap in the original, now-cut gene by splicing the open ends together, resulting in a deletion. There are many potential issues with type II CRISPR-Cas9 systems in the context of gene editing, namely via potential for unwanted off-target binding stemming from the single guided nature of the system and / or NHEJ, which will be discussed later.
Cytosine/Adenine Base Editors
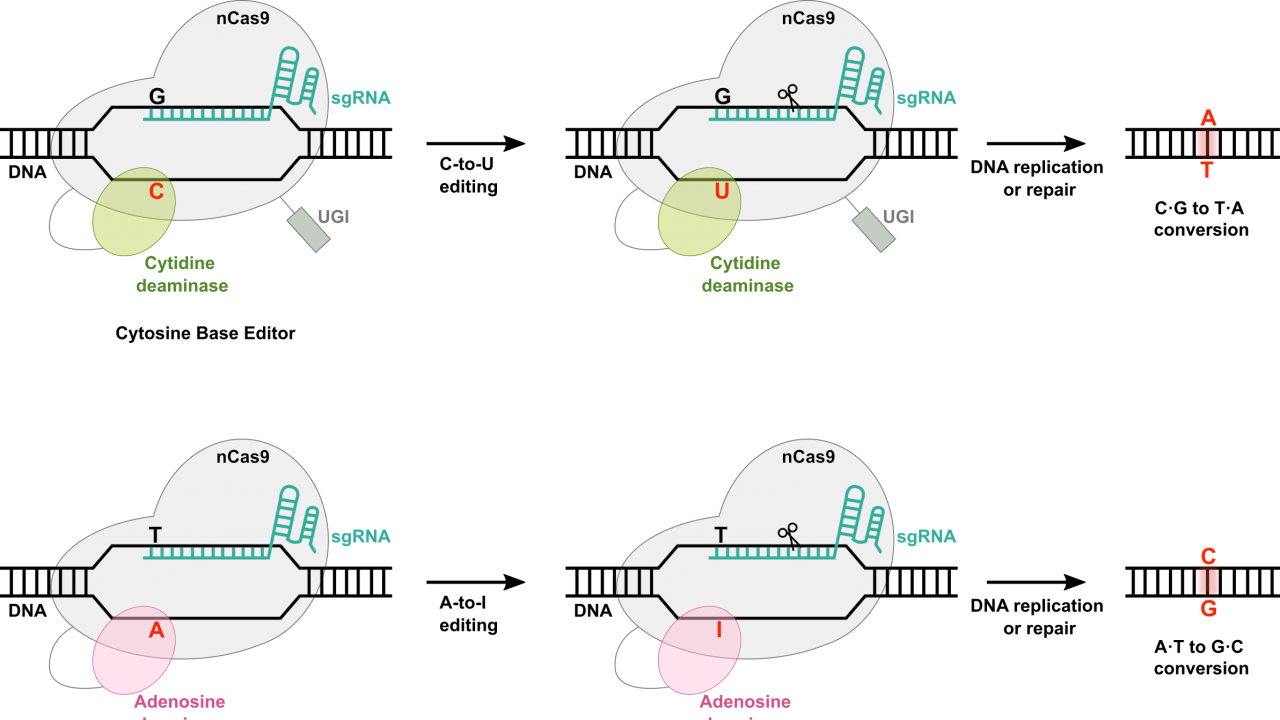
Figure source: UC Davis Biotechnology program
At the most basic level, base editing enables point mutations through the direct conversion of one DNA base pair to another, addressing some limitations associated with CRISPR-Cas9 systems. Much of the machinery remains the same for base editors as in other Cas9-dependent systems. Included in every base editor is a catalytically impaired Cas9 nickase or Cas9n (also known as dead Cas9 – great band name potential – or dCas9), a single guide RNA (sgRNA) complementary to a target region in the host genome, and a base editing enzyme, such as a cytidine deaminase or an adenosine deaminase that allows for base swaps.
As with all other Cas systems, the host-complementary sgRNA is designed to recognize and direct the base editing machinery to the target site within the genome. Once at the target site, the Cas9n binds to the DNA, though this time, it does not introduce a break of any kind. Instead, the base editing deaminase, which is fused to the Cas9n, facilitates the direct conversion of a specific base pair. For example, cytidine deaminases can convert cytosine (C) to uracil (U), which is then read as thymine (T) by the cellular machinery, while adenosine deaminases convert adenine (A) to inosine (I), which is read as guanine (G).
Base editing thus allows for targeted single base pair substitutions without introducing double-stranded breaks or relying on the cellular repair pathways, reducing the likelihood of unintended insertions or deletions. Though this might seem like a rather minor advancement for the field, prime editors actually present significant advantages when considering the challenge of correcting the nearly-80% of human genetic diseases caused by point mutations. Additionally, a number of target genes are just a point mutation away from enhanced functionality (like increased muscle mass via myostatin inhibition or Alzheimer’s resistance stemming from single amino acid change in ApoE).
Each of these technologies offers its unique advantages and challenges for researchers and clinicians. By understanding the mechanisms of action and optimizing the design of these gene editing tools, scientists can enhance their specificity and reduce the likelihood of off-target effects. In practice, the latter 3 – all Cas-based – dominate the editing landscape, and are what we focus on throughout the rest of the article.
Prime Editing
Figure source: Wikipedia
A novel gene editing technique, prime editing provides a refined approach for making precise alterations to DNA sequences, circumventing some of the issues associated with CRISPR-Cas9 systems. The prime editing system consists of three key elements: a prime editing guide RNA (pegRNA) complementary to a region in the host genome and includes the RNA template to be inserted, an impaired Cas9 nickase (nCas9) incapable of double-stranded breaks, and Moloney Murine Leukemia Virus reverse transcription enzyme (M-MLV RT, abbreviated as RT going forward).
Similar to traditional gRNAs, pegRNA is designed to recognize and guide the prime editing machinery to the desired target site within the genome. Upon reaching the target site, the nCas9 enzyme creates a single-stranded break, or "nick," in the DNA. The RT enzyme then synthesizes a new DNA strand, using the pegRNA as a template containing the intended edit. The modified strand is subsequently incorporated into the host genome to “fill the gap” created by the nickase, with the final step being the replacement of the now-mismatched original host genome segment via cellular mismatch repair. Unlike CRISPR-Cas9, prime editing allows for targeted insertions, deletions, and, uniquely, single base substitutions (more on this later), thereby allowing a much more diverse array of allowable edit types. Areas for improvement of prime editor complexes mainly revolve around improving editing efficiency and optimization of pegRNA design.
The Challenges in Gene Editing and Possible Solutions
Off-target edits
The biggest hurdle facing Cas editing systems is the risk of off-target edits – unintentional edits to additional or alternative bases, resulting in deleterious edits in untargeted regions of the genome. These accidental alterations can have harmful consequences, in particular the activation of oncogenes that trigger cancer. As such, researchers are tirelessly working to improve the specificity of Cas editing systems to minimize the occurrence of off-target edits. Right now, a lot of progress is being made but inconsistent evaluations and experimental conditions make assessing the state of things and ensuring progress builds on itself difficult.
How high is off-target edit risk for existing editors?
While all Cas editors come with some risk of off-target edits, the actual rate of occurrence relative to natural cell-mediated processes (see this Appendix section for some loose estimates of the natural mutation rate) is unknown. If we knew this rate, it could act as a useful benchmark to potentially increase our confidence in editor(s). Even in vitro, knowing that an editor had an off-target edit rate substantially lower than that of cell division even in vitro (and doesn’t preferentially edit oncogenes or other harmful regions) would provide a helpful complement to trials and animal studies alone.
How do different editing systems stack up in terms of off-target edit risk?
Given this uncertainty, it’s hard to get a precise answer to the question of which editor has the lowest off-target edit rate. As best as we can tell, mainly according to this paper, the most correct ordering from lowest to highest is:
Prime editor: Prime editors have the lowest off-target edit rates of the three technologies.
Base editor: Base editors have a higher off-target edit rate than prime editors, but they are still more precise than CRISPR-Cas9.
CRISPR-Cas9: CRISPR-Cas9 has the highest off-target edit rate of the three technologies.
Again though, there are tons of papers discussing this so these are very tentative general rankings. It’s also important to remember that the off-target edit rate of each technology can vary depending on the specific application and that these rankings are coming from the “standard” version of each of these technologies.
Types of off-target edits
Cas editing systems’ off-target effects fall into two broad categories: “distal” and “proximal” (source of terminology). An edit is proximal if it falls <200bp from the intended target site and distal if it falls outside that window.
Distal edits have two sub-categories: guide-dependent and guide-independent off-target edits. Guide-dependent off-target edits result from off-target binding of a guide RNA (gRNA) to the wrong region, either because of an exact complementarity or, more commonly, a close enough but not exact complementarity to the guide.
Proximal edits occur within the editing window and include:
Bystander edits: additional or alternative unintended mutation to a base near the target. One of the more common off-target edit types for cytosine/adenine base editors.
Deletions caused by NHEJ: as mentioned already, CRISPR-Cas9’s double-strand breaks come with the risk of the two cut ends joining directly rather than the desired homology-directed repair.
Strategies for reducing off-target edits
Off-target gRNA binding is a problem for all three of CRISPR-Cas9, prime editing, and cytosine/adenine base editors, meaning gRNA design is an especially valuable topic to work on. Progress today primarily involves designing guides that are less likely to accidentally bind to an off-target region of the genome and will edit the target region with higher efficiency. There are different approaches to this but because it’s ultimately a sequence design problem, we are most excited about approaches that leverage ML and large datasets. Early methods that design guide RNAs like DeepCRISPR, Microsoft Research’s Azimuth/Elevation, and, more recently, DeepPrime show promising progress towards being able to predict off-target binding. However, these papers only evaluate prediction of known off-target sites in vitro so we don’t yet understand how much of a benefit they deliver in a clinical setting. That said, like many areas where more data is being generated at lower costs, we can expect methods to continue to improve here.
Engineering the Cas protein for higher sequence specificity also benefits all three Cas-based systems and has the potential to reduce both distal and proximal off-target edits. In this and follow-up work, Kim et al. designed (the extremely cool-ly named) Sniper family of Cas9 variants which maintain on-target efficiency while reducing off-target double-strand breaks via a clever directed evolution scheme in E. coli. In an unbiased genome-wide screen, Multiple of the Sniper Cas9 variants cleaved far fewer sites than WT Cas9. Furthermore, when paired with BE3, a base editor, Sniper-Cas9 had a 2-16 fold lower off-target edit rate than the wild type. The fact that editing Cas9 enabled gains for both CRISPR and CBE/ABE specificity suggests that improvements to the Cas protein may generalize across editing systems, which would make Cas engineering even more valuable. In the future, we expect ML-guided protein design to help design even better, more specific Cas9 variants. We’re also optimistic that gains made towards editing vanilla Cas9 will be translatable to dCas9 and nCas9.
Because CRISPR-Cas9 is increasingly being replaced by CBEs/ABEs and prime editing, most of the work we are aware of on reducing bystander edits focuses on CBEs/ABEs. Multiple papers have reported on variants of one or both editors with engineered deaminases that maintain or improve on-target efficiency while lowering off-target deaminase activity via directed evolution (TadCBE) or rational protein engineering of the deaminase. This sometimes comes with the additional benefit of reduced off-target distal activity. Both this paper, which replaced the commonly used cytidine deaminase in CBEs with a more sequence specific one, and TadCBE found that improved specificity also reduced distal off-target effects.
Stepping back, while off-target effects remain a major problem for editing systems, we’re confident that the combined progress from improving guide design, the Cas protein, and system-specific components can combine to drive rapid progress towards the goal of negligible off-target effects. Rather than progress along individual axes, we see lack of comprehensive datasets and measurement experiments as bigger obstacles towards actually translating off-target effect mitigations to clinical settings. In What’s Next?, we propose some ideas for eliminating this bottleneck.
Immunogenicity of Editors
Another challenge specific to Cas editing systems is immunogenicity, which occurs when a therapeutic agent is blocked by the host immune system by clearance or neutralization – distinct from immunogenicity to viral vectors (covered earlier). Because the most common Cas orthologs stem from S. aureus and S. pyogenes, the dominant concern when thinking about immunogenicity is a response that comes from natural exposure to bacterial proteins at some point prior to therapeutic delivery. Though the specific numbers differ across studies, it’s clear that many individuals possess natural antibodies to SpCas9, the enzyme utilized in CRISPR-Cas9 gene editing, with occurrence of anti-Cas antibodies at potentially as high as 78% to 95%. When the Cas9 protein or RNA is introduced into the body, these pre-existing antibodies may provoke an immune response specific to editing elements themselves that can complicate and potentially limit the ability to perform subsequent edits. Evidence in preclinical studies demonstrates that immune response to Cas9 is a legitimate concern, though it is unclear how vector-independent immunogenicity will influence therapeutic efficacy.
If adverse reactions to cutting enzymes themselves pose a risk to treatments there are fortunately a few ways to make a less immunogenic Cas. The first, least palatable option – which is possible now – is to simply serially undergo multiple treatments that depend on different CRISPR effector proteins. This would look like round 1 of treatment relying on S. aureus-derived Cas, while round 2 uses C. jejuni, as antibody-specific responses are not cross reactive between species. Though doable today, this solution is not ideal due to the reliance on naturally occurring enzymes, especially if the goal is to achieve either concurrent dosing of different therapies, or many sequential rounds of treatments for different targets. For the orthogonal effectors that do have some overlap, use of immunosuppresants could allow for more tolerable intake of therapies, though that comes with its own challenges.
Engineering of various CRISPR effector proteins remains the best approach to solving the immunogenicity issue. One approach involves engineering Cas proteins to generate more orthologs that can be serially or concurrently delivered to the same patient. Though much of the work in machine guided design for enzymatic editors revolves around improving cutting efficiency and specificity, much of the same principles could be applied to design around motifs involved in immune response. In the same vein, another promising strategy involves rational design strategies to mask specific epitopes to prevent immune responses. A group in China managed to avoid toxicity from the Cas protein itself by first identifying problematic sources of immune responses to Cas9, then engineering the enzyme with a simple R338G substitution to avoid B cell immunogenicity.
Again, relative to negative immune responses to delivery vectors themselves, which is certainly a real and known issue in the field, it really is not known how critical immunogencity concerns are for successful repeatable edits, but it’s worth covering options once the field develops a clearer picture for the problem (more on that later in What’s Next?).
Universality
Lastly, a more general challenge in the field of gene editing is developing techniques that work efficiently and flexibly across the entire human genome. Whether in its inactive base editor form or vanilla CRISPR-Cas9 mode, the reliance on PAM recognition as part of the Cas mechanism acts as a potential barrier to target selection, since it limits gRNA binding to locations that have a suitable PAM sequence the right distance from the target itself. The good news is help is definitely on the way to solve this problem! Academics have made great strides to create more flexibility by engineering CRISPR-Cas9 nucleases with altered PAM specificities, and in some cases, have even generated near-PAMless variants, allowing for unconstrained target selection. As far as issues go with editing, this one seems the closest to being fully solved. Great job, everyone!
Appendix
Establishing a natural mutation rate baseline
Helpful numbers / estimates
Number of cells in a human body: 4e13
Number of DNA damage events per cell per day (source): >10,000
Number of somatic mutations per year for different tissue types (source):
Mutation rate per base pair per generation in humans (source): 2.5e-8
Approximate number of base pairs in the human genome: 3e9
Number of mutations per cell division in humans (estimate): ~0.1-10
Mutation rate of DNA polymerase…
Another source gives a direct estimate of 1.14 mutations per cell division in hematopoiesis
Number of cell divisions in a human per day (source): 3e11
Note: This is actually off because it’s averaging over a lifetime but in practice the rate will be much higher during development.
Loose bounds on acceptable number of mutations from editors
Let’s take as an assumption that if our off-target edit rate is on average below the ongoing mutation rate of normal body processes than we’d be happy. Using our number of 3e11 cell divisions in a human per day and 1 mutation per cell division, we get a very hand-wavey estimate of 3e11 mutations per day as part of ongoing biological processes. If we assume an edit we want to apply will edit a good fraction of all cells (a very conservative assumption), then this suggests we want to keep our off-target rate below 1e-2 in order to match the total number of mutations from a day of cell divisions.
Alternatively, if we just compare editing to a cell division event directly, we could argue that the per cell off-target rate should be at or below 0.1 mutations per cell.
Caveats
There are lots of reasons why this estimate is naive and doesn’t actually work. It should be taken only as a very loose guess to hopefully be supplanted by Longitudinal / observational studies in model systems in the future. To name just a few of these reasons:
Most editing interventions won’t actually aim to edit all human cells.
Mutation rates differ between parts of the genome and type of mutation and are normally under selection pressure. This means that mutations due to off-target edits can be a difference in kind rather than just in number. For example, off-target edits in oncogenes are much worse than a randomly chosen mutation so interventions that preferentially incur those are more dangerous than their raw rate would suggest. On the flip side, this also means we can directly check for them.
Some of these numbers may not account for DNA repair machinery which protects against dangerous mutations during DNA copying but may not work as well for off-target edits.
Thanks to Milan Cvitkovic, Willy Chertman and Neal Nolan for help reviewing this piece!